The width of an individual DNA strand is around 2.5 nanometers. The smallest transistors that we can produce today are around 3 nanometers wide. Although we are already capable of manufacturing some devices at levels of precision rivaling the cell, there’s plenty of room for more innovation. In a landmark lecture given by Richard Feynman at Caltech in 1959, he discusses the theoretical limits of manufacturing and notes that the possibilities really go down to the level of manipulating individual atoms to create the structures and devices we want.
Feynman didn’t get too carried away with speculation, but he made it clear that the world would look very different if we one day attained the ability to construct arbitrary structures atom by atom. The talk preceded the launch of shows like Star Trek, which dazzled imaginations with machines like the replicator, capable of synthesizing meals — and martinis — on demand. Neal Stephenson’s Diamond Age later envisioned a world where households replaced garbage cans with ‘matter compilers’ which recycled waste into its constituent atoms to recycle them for future use.
Of course, atomic or molecular scale manufacturing wouldn’t just revolutionize domestic life, it would have consequences across all domains of industry with massive implications for chemistry, material science, construction, medicine, and biology writ large. Two prominent thinkers exploring molecular machinery, J. Storrs Hall and Robert Freitas, estimated that “mature nanotechnology could replicate the entire capital stock of the United States – ‘every single building, factory, highway, railroad, bridge, airplane, train, automobile, truck, and ship’ – in a mere week.”
To be clear, the ability to conduct precision manufacturing at the atomic level is still a prospect that is, at the least, decades away. We might not arrive at generalized atomic-scale manufacturing any time soon, but it is becoming feasible to perform sophisticated industrial processes and build interesting systems on the scale of larger molecules. With the introduction of artificial intelligence technologies capable of designing and anticipating protein structures, the near-term prospect for synthesized molecules in industry is immense indeed. To examine how this kind of frontier research may have broad implications for vast sectors of our economy and society, we’ll journey into the technical and biological bases for these ideas and explore how they are being tested and implemented today.
A Brief History of Atomic-Scale Thinking
In his 1959 lecture on the subject, Richard Feynman didn’t get too deep on specifics of how exactly tiny molecular machines could be produced, though he did provide one intriguing conceptual scheme. Today, human hands can be used to control much smaller robotic hands, such as the da Vinci surgical robot. Feynman envisioned a cascading system of such robotic hands, each one smaller than the next, manipulating ever smaller components, until finally the smallest set of robot hands is building gears and motors made of individual atoms.
Since Feynman’s talk, many conceptual models of atom-sized machine parts have been imagined and designed; like the molecular logic gate, devised at IBM in 1988, or the synthetic molecular motor, designed in 1999.
Source: Molecular Motors, 2014
Throughout the 1980s, the first attempts to actually synthesize some of these atom-sized machine parts took place. However, the progress was slow and incremental. As Dexter Johnson, a molecular nanotechnology writer, put it:
“First, in 1983, [Jean-Pierre] Sauvage was able to link two ring-shaped molecules to form a chain. Then, eight years later, [Sir J. Fraser] Stoddart, a professor at Northwestern University in Evanston, Ill., demonstrated that a molecular ring could turn on a thin molecular axle. Then, eight years after that, [Bernard] Feringa, a professor at the University of Groningen, in the Netherlands, built on Stoddardt’s work and fabricated a molecular rotor blade that could spin continually in the same direction.”
On the back of this research, in 2005, a Dutch professor from the University of Groningen named Bernard Feringa even demonstrated how it would be possible to construct a nano-scale car propelled by chemical energy. In 2016, Feringa and his collaborators Jean-Pierre Sauvage and Sir James Fraser Stoddart were awarded the Nobel Prize in Chemistry for their achievements. However, synthesizing nano-scale objects like the car’s molecular motors and chassis remains to this day a piecemeal operation. These molecular machines were manufactured from organic molecular compounds rather than in an atom-by-atom fashion.
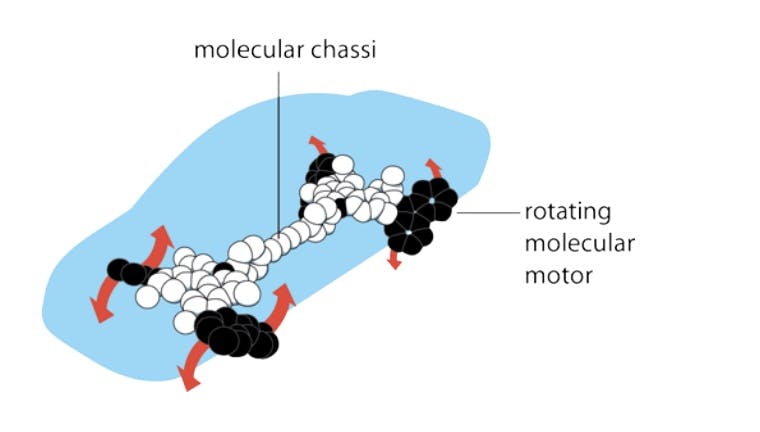
Source: Nobel Prize, 2016
Eric Drexler, a pioneering thinker in the field of molecular manufacturing, said in 2022 that we’re still only “in the process of building tools that will enable us to build tools that will enable us to build tools that will allow us to move in that domain.” Furthermore, even if we were able to build molecular gears, motors, and rotors at scale, it is unclear what we could use them for, or if they would necessarily even need to take on these form factors in the first place.
However, in a seminal paper on molecular manufacturing in 1981, Drexler pointed out that perhaps we don’t necessarily need to create all these molecular components de novo, or atom-by-atom. After all, the organic molecules ruling all living organisms, namely proteins, already perform machine-like roles. For instance, the actin and myosin proteins in our muscles act like actuators. Collagen acts like a cable, capable of transmitting tension. Enzymatic binding sites operate like clamps, holding workpieces in place, and so forth. In other words, perhaps we need not reinvent the wheel. The hardware for assembling molecular machines already exists — it’s in our cells!
Bridging the Gap with Biology
How does a seed become a plant, or a baby a full-grown adult? Molecular machines are the answer. All the compounds in a biological organism are made of just a handful of kinds of atoms, consisting mostly of carbon, oxygen, hydrogen, nitrogen, phosphorus, and a few others.
Some of the most important compounds that these atoms are arranged into are proteins. This is because proteins are the most versatile organic compound. Just as Legos can be used to build up a large variety of structures and machines from simple constituent pieces, proteins can form nearly any arbitrary structures and machines in the molecular world.
For example, proteins can be arranged into enzymes, which facilitate everything from digestion to metabolism and DNA replication. They can be used for structural support, forming collagen which creates fibrous networks for holding tissues together. They can be used to carry oxygen molecules in the bloodstream, create antibodies that defend an organism from antigens, form the hormones that transmit messages to regulate physiological processes, and even accrete together to become the very stuff that powers our muscles.
Each cell in the human body has a protein production site called the ribosome. Ribosomes produce proteins using a standardized method. Every protein is a chain of amino acids. There are only 20 amino acids that proteins can possibly be made of, although they can consist of hundreds or thousands of amino acids strung together. The order of the amino acids in the chain determines the ultimate shape that the protein will take on, and therefore, the function that protein will eventually have.
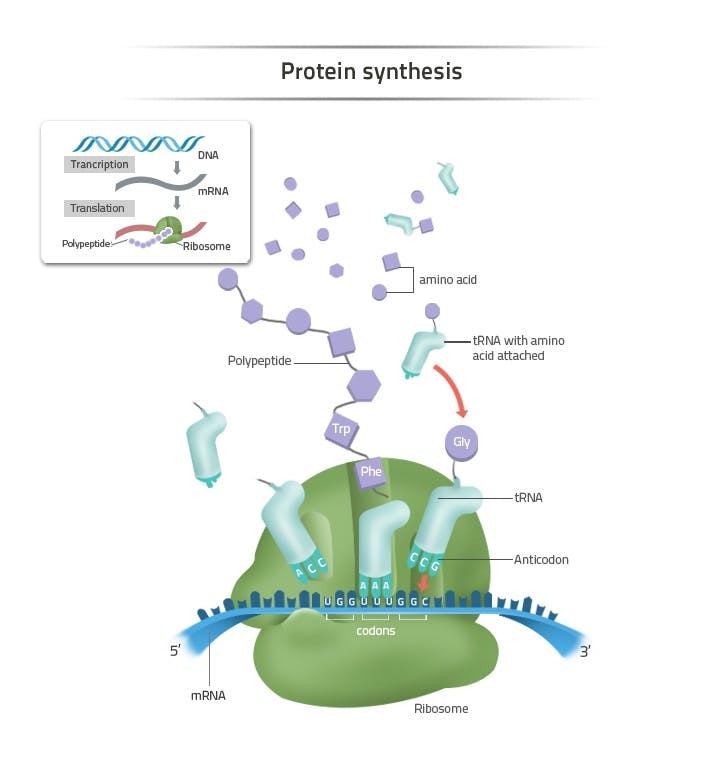
Source: Labster
The correct ordering of amino acids required for protein construction is stored in DNA. In fact, that’s really all that DNA is – a memory base for protein assembly. Each gene encodes the sequence for a particular protein. To produce proteins, mRNA copies over the protein recipe from DNA found in the nucleus of a cell and transports it to the site of the ribosome, a literal industrial complex surrounding the locus of DNA storage.
Using Proteins to Engineer Molecules
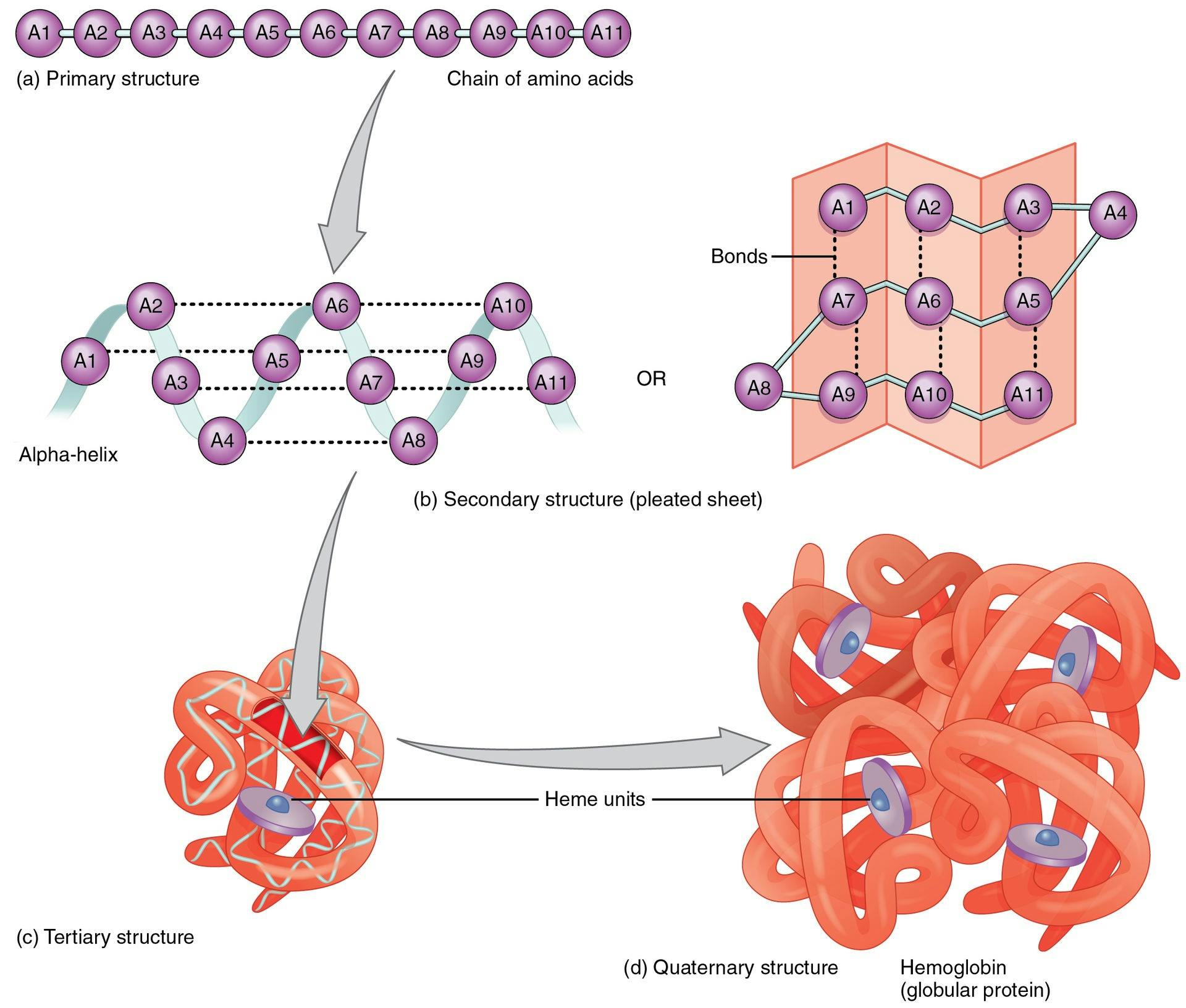
Source: Openstax
In other words, the hardware for molecular machinery already exists with the ribosome, and even the programming language required already exists in the form of DNA. Turns out all we need to do is learn to write programs, in effect commanding the DNA to create the proteins we want.
Doing this DNA programming has been historically very difficult. For a long time, when given some sequence of amino acids, it was nearly impossible for humans to figure out how exactly that chain of amino acids would fold, and therefore what structure that protein would ultimately take on. There were just too many possible interactions between the constituent atoms. So, while the structure of shorter chains was easier to ascertain, it would have been mind-boggling to evaluate how a 500-long amino acid chain would shape up.
All this changed, however, with the maturing of artificial intelligence. In 2018, DeepMind released AlphaFold, an artificial intelligence program capable of predicting protein structure from amino acid sequences with a high degree of accuracy. In 2020, AlphaFold 2 was released with increased the program’s accuracy further still.
Advances in artificial intelligence are now also showing massive implications for protein design. Large language models, it turns out, aren’t just good for writing poems in iambic pentameter, they’re now fully adept at designing proteins capable of performing specific tasks. In a few recorded cases, the enzymes designed by artificial intelligence have proven even more efficient at their tasks than enzymes found in nature.
The language of genetic code enables the production of proteins, so gaining an intimate knowledge of reading and writing that genetic code is required for better protein design and synthesis. It’s rather lucky for molecular engineers that the cost of sequencing DNA has fallen dramatically over the previous two decades. Genetic synthesis, or the ability to produce artificial genetic material, has also fallen in price over the previous two decades, though not as drastically as the cost of sequencing.
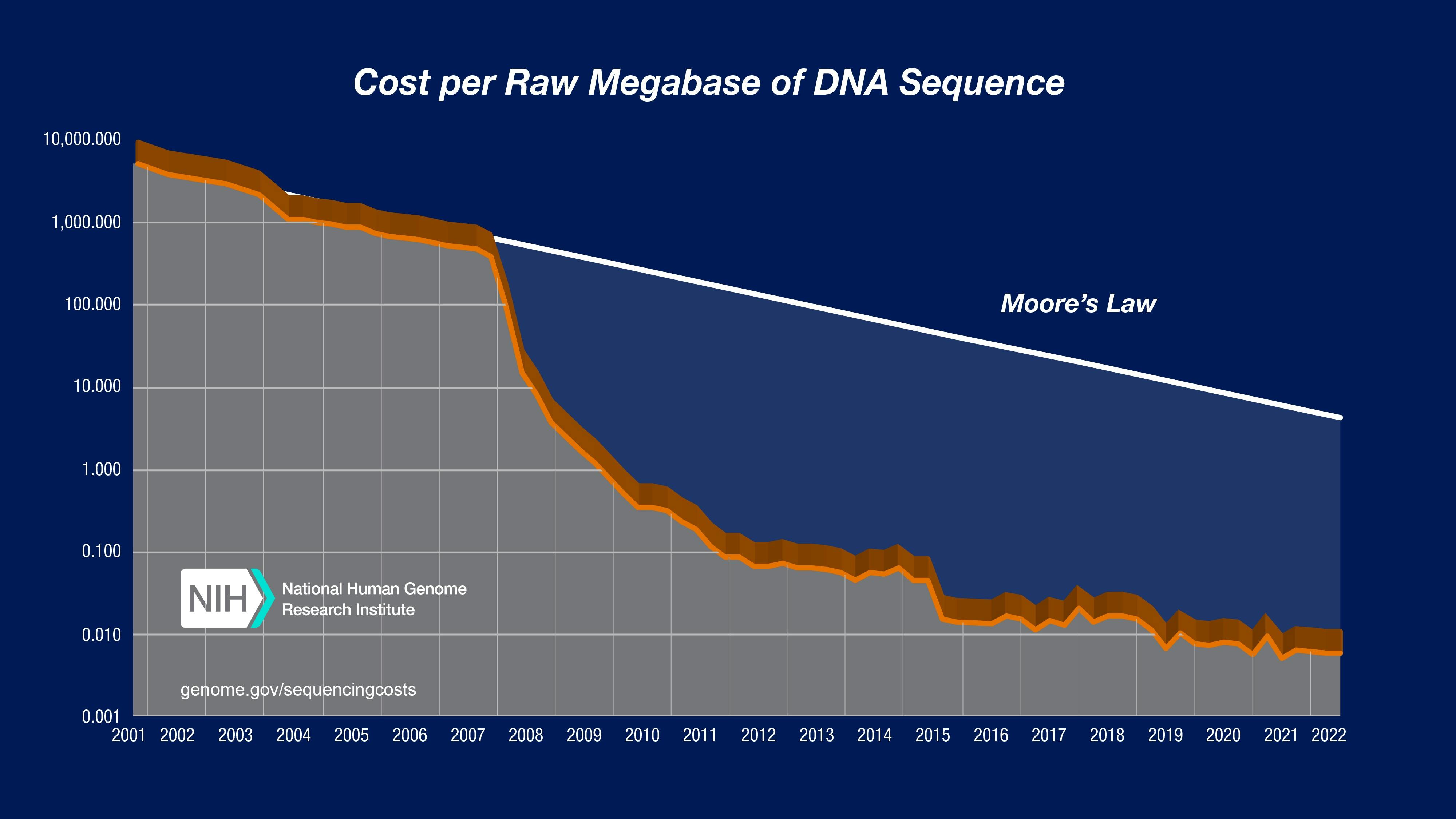
Source: National Human Genome Research Institute
Our increasing understanding of genetics points to a future where the hardware of biology may become a very useful paradigm for manufacturing new industrial products. In fact, this is exactly what got Eric Drexler so excited about proteins and the ribosome in the first place.
As Eli Dourado puts it, “once the designer has determined the desired [amino acid] order, she could synthesize the protein on a tabletop device or encode it as genetic material and give it to a living ribosome (this is what the mRNA vaccines did). Ribosomes produce proteins much faster than state-of-the-art lab equipment – human ribosomes can bond about two amino acids per second (and bacterial ribosomes are yet faster), while tabletop reactions happen at a rate of one bond every 2.5 minutes. This superior performance in an apples-to-apples task is just one indication of how much more efficient molecular machinery can be compared to bulk chemistry.”
What exactly are the applications of such designer proteins? One answer to this question is already evident in mRNA vaccines, which were first approved for wide-scale use during the COVID-19 pandemic. mRNA vaccines code for newly designed antibodies that our cells are instructed to produce. Synthetic proteins can be engineered to better encapsulate drugs and deliver them more precisely to specific targets in the body. They can also create scaffolds for tissue engineering. Another fascinating application of designer proteins is bioremediation. Engineered proteins can be designed to degrade or transform pollutants, such as toxins or pollutants in water or soil, helping to clean up contaminated environments.
Perhaps the most economically salient application would be the use of synthetic enzymes as biocatalysts. This could improve the efficiency of chemical reactions, which could help in the production of fuels, pharmaceuticals, chemicals, and materials of all kinds. In the realm of carbon capture and utilization, for instance, many see a future where carbon captured from the atmosphere could be recycled into a host of useful materials from plastics to fuels. The bottleneck to doing this now is the energy involved in getting carbon to react such that it can form complex molecular structures. It is hoped that in the near future, designer proteins and intricately designed molecular machines could be used to accelerate these processes and decrease the amount of energy required to effectively turn captured carbon into useful materials.
The Route to Atomically Precise Manufacturing
Though the future for bio-industry seems bright, many must still be wondering about the prospects for nano-scale technology. Which efforts are being undertaken to enable atomically precise manufacturing? What are the barriers that still lay before us in that domain?
First off, let’s lay the groundwork for what kind of manufacturing is possible today on a nano-scale. Photolithography and electron beam lithography, used in the production of microchips, are techniques for manipulating materials at a nanoscale. Both involve removing material rather than depositing it.
In a semiconductor fab, photolithography techniques are conducted on two axes. In 2021, research emerged out of Purdue showing that atomically-precise etching on three dimensions is now possible using precise photolithography. With this technique, it’s possible to create smooth and even rather intricate nanostructures, although they are still fundamentally carvings produced out of a fixed pre-existing material.
Other techniques like atomic layer deposition and molecular beam epitaxy, which can “grow” materials layer-by-layer, can produce material films with atomic-scale precision. Both are also often used in the world of semiconductor fabs. However, they still approach nanomanufacturing at a bulk level, applying atoms in layers rather than one by one. Furthermore, they only work with certain kinds of molecules. The ultimate goal of nano-scale manufacturing is to arrive at a process that is capable of assembling molecular machines atom by atom.
At present, the technology closest to accomplishing this is a scanning tunneling microscope (STM). STMs possess extremely sharp tips with edges that are only a few atoms wide. The microscope works by getting its tip so close to the sample material that the electrons from the sample material start to “tunnel” or teleport through the tiny gaps between the tip and the sample. Then, by measuring the rate of electron tunneling at different points across the sample, the STM can determine with extremely high precision the topography of the sample. In 2003, scientists used a scanning tunneling microscope with a monoatomic tip to make and break the covalent bonds in a layer of silicon, essentially producing the first demonstration of a technique that can add or remove individual atoms from a material.
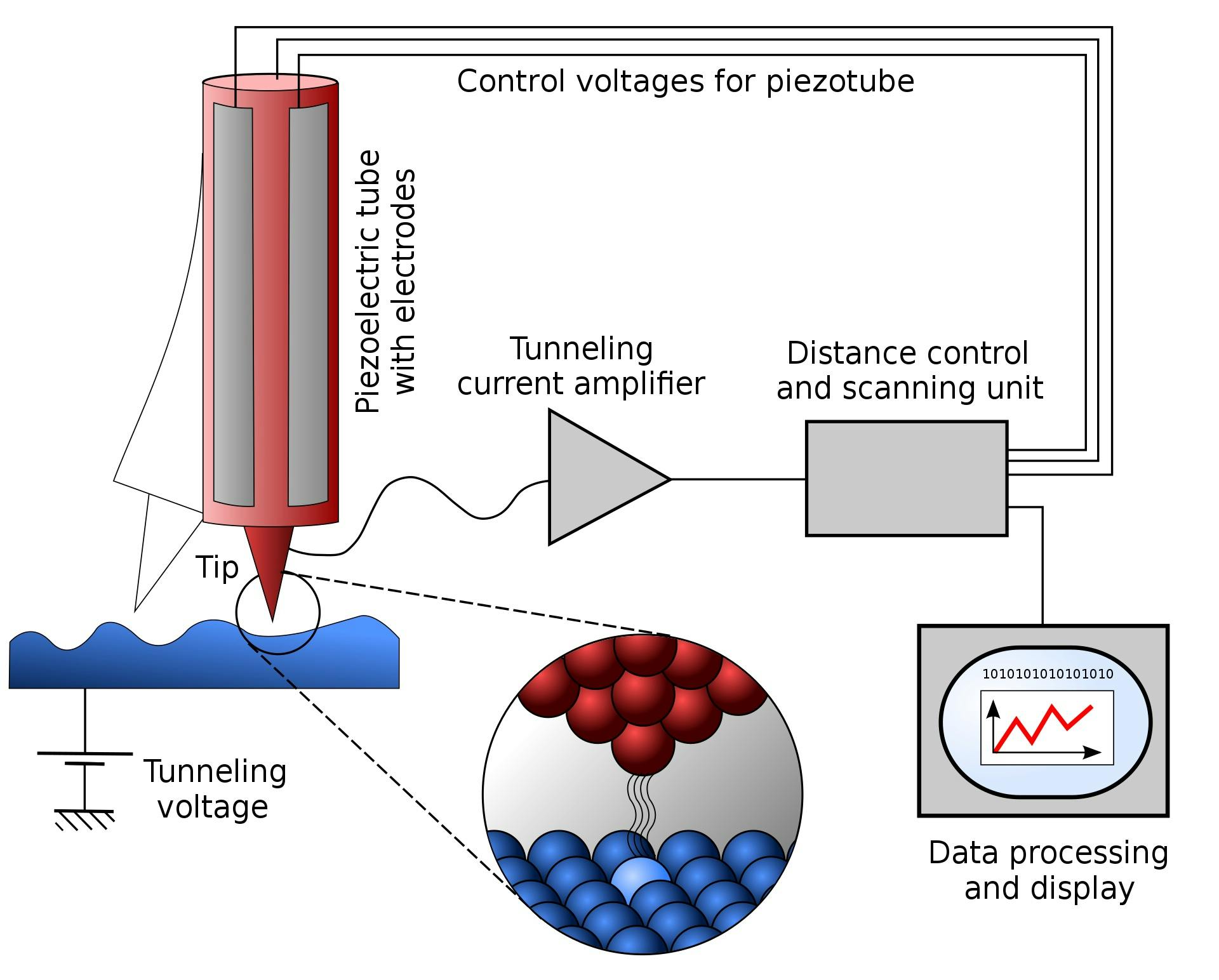
Source: Michael Schmid and Grzegorz Pietrzak
Though this was an impressive achievement, true atomic-scale manufacturing would require doing this with many different STMs operating simultaneously. After all, even objects as small as an apple have billions upon billions of atoms that would need to be precisely placed by such a machine.
Synthesizing atomically precise components is one thing. Even assuming that it would be possible to build tiny molecular machine gears and motors atom-by-atom, it’s another challenge entirely to figure out how exactly all these components would interact in a complex system or in which medium they would even interact in the first place.
To illustrate this point, the best atomically-precise molecular machinery we know of today, the ribosomes in our cells, conduct their operations in water. However, suspending molecular machinery in a solution is not the best way to produce a reliable system. Molecules suspended in liquid experience frequent and disruptive collisions with the fluid particles due to Brownian motion.
To get a machine to work repeatedly and reliably, the components need to be fixed together, or held in place, in some way or another. A method of producing reliable interactions between components so tiny has not yet been designed. However, some are exploring hypothetical alternatives like using graphene or other crystalline structures as the mediums where such molecular interactions can happen. Crystalline structures are solid, so they are held in place, yet there are very large gaps between the atoms, which allow molecular machines to be integrated into the crystals and still have room to pass inputs back and forth.
The technical challenges until Feynman-style universal molecular assemblers become feasible are, to put it mildly, gargantuan. However, even if we will not be able to build replicators or matter recyclers any time soon, the prospects already on the horizon with the maturing of DNA and protein synthesis are immense. There might soon even be the possibility of merging findings on the frontiers of atomic material manipulation with those of protein design and synthesis, as suggested in a talk by Jacob Sweet called “There’s Plenty of Room in the Middle.”
In the early 2000s, the aspirations for nanotechnology were extremely ambitious, as evidenced by Ray Kurzweil’s prediction in 2000 that “Around 2030, we should be able to flood our brains with nanobots that can be turned off and on and which would function as ‘experience beamers’ allowing us to experience the full range of other people’s sensory experiences…Nanobots will also expand human intelligence by factors of thousands or millions. By 2030, nonbiological thinking will be trillions of times more powerful than biological thinking.”
Though Kurzweil’s timeline was overly optimistic, the excitement surrounding the potential of nanotechnology has certainly captured the imaginations of many ambitious scientists and researchers. On the back of the work of Eric Drexler, new and exciting ventures have sprung up to support it. One such group is Speculative Technologies, helmed by Ben Reinhardt. Among other things, Ben is keen on encouraging researchers to think more expansively and intuitively about how tiny nano-molecular components could work together in complex systems. To this end, the organization has procured funding to support efforts like a molecular additive manufacturing program and a nanomodular electronics program.
Although we might not yet have entered the age of true nano-manufacturing, such examples point to a renewed interest and energy around nanotech research that is laying the groundwork for future breakthroughs. Given the nearly unlimited potential of nano-scale molecular manufacturing and the already rapid pace of progress, the era of replicators and matter compilers may not be too far off.
Disclosure: Nothing presented within this article is intended to constitute legal, business, investment or tax advice, and under no circumstances should any information provided herein be used or considered as an offer to sell or a solicitation of an offer to buy an interest in any investment fund managed by Contrary LLC (“Contrary”) nor does such information constitute an offer to provide investment advisory services. Information provided reflects Contrary’s views as of a time, whereby such views are subject to change at any point and Contrary shall not be obligated to provide notice of any change. Companies mentioned in this article may be a representative sample of portfolio companies in which Contrary has invested in which the author believes such companies fit the objective criteria stated in commentary, which do not reflect all investments made by Contrary. No assumptions should be made that investments listed above were or will be profitable. Due to various risks and uncertainties, actual events, results or the actual experience may differ materially from those reflected or contemplated in these statements. Nothing contained in this article may be relied upon as a guarantee or assurance as to the future success of any particular company. Past performance is not indicative of future results. A list of investments made by Contrary (excluding investments for which the issuer has not provided permission for Contrary to disclose publicly, Fund of Fund investments and investments in which total invested capital is no more than $50,000) is available at www.contrary.com/investments.
Certain information contained in here has been obtained from third-party sources, including from portfolio companies of funds managed by Contrary. While taken from sources believed to be reliable, Contrary has not independently verified such information and makes no representations about the enduring accuracy of the information or its appropriateness for a given situation. Charts and graphs provided within are for informational purposes solely and should not be relied upon when making any investment decision. Please see www.contrary.com/legal for additional important information.