Biology is kaleidoscopically complex. We are still far from understanding how all the components, even at the level of a single cell, work together — let alone at the level of an entire organism. For a long time, scientists had to rely on imperfect and limited methods of observation to build an understanding of biology.
To the credit of previous generations of biologists, this got us pretty far. We discovered the existence of organelles, genetic material, and many other mechanisms that drive a number of critical biological functions. However, for a long time, it was as though we could only investigate biology from a great distance, like trying to understand the inner workings of an organization by peering through its windows.
This all changed with the discovery of CRISPR technologies in 2012, which granted researchers an unprecedented degree of control over genetic material. For the first time, it was possible to not only observe biology, but perturb it, alter it, and even design it.
To some extent, humans already have a storied legacy of tinkering with genetic material. For millennia, these practices have largely been confined to breeding schemes resulting from agriculture and domestication. Nearly all the produce we eat on a daily basis, for instance, is the product of hundreds, if not thousands, of years of selective breeding.
Many might be interested to learn, for instance, that broccoli, cabbage, cauliflower, kale, and collard greens are in fact all the same genetic species, just bred over generations to express different traits.
Of course, such breeding techniques took generations to achieve the desired results. With today’s CRISPR technologies, genetic modifications can be made directly, allowing us to eliminate or insert individual genes – even individual base pairs. This level of precision has produced a revolution in biology, allowing us to plumb the inner workings of cells and genes just as one might uncover the inner workings of a gadget by taking it apart to understand the function of each constituent component.
This new set of tools and abilities has unlocked tremendous opportunity, with untold applications within the realms of therapeutics, industry, and research. In just a few short years, CRISPR has moved the field of biology outside of the realm of mere observation and analysis and placed it squarely within the domain of engineering.
A New Biological Paradigm
In reality, it’s not merely CRISPR technology that has allowed us to begin performing precision genetic engineering. A number of new technologies that matured in the early 21st century have operated in concert to produce our ability to perform unthinkable biological interventions.
Among these are DNA sequencing technologies, which allow us to read the base sequences of specific DNA fragments or entire genomes; DNA synthesis technologies, which allow us to print desired strands of DNA; and finally, gene editing technologies like the suite of CRISPR tools, which allow us to delete or insert genetic material with a high degree of precision.
Taken together, these techniques constitute the pillars of an entirely new branch of biology called synthetic biology. Synthetic biology is concerned with exploring the range of possibilities opened up by genome editing technologies, including the possibilities of re-engineering existing organisms or designing entirely new ones. The ethos of engineering and design fully permeates this new discipline, a fact perhaps illustrated mostly clearly by synthetic biology’s absorption of engineering vocabulary. A host organism, for example, is called a “chassis,” whereas engineered genes are “genetic circuits.”
Alexandra Ginsberg, an artist exploring the role of aesthetics in synthetic biology, has spent years contemplating the field’s implications for our understanding of nature as such. “How will we classify what is natural or unnatural when life is built from scratch? Synthetic biology is turning to the living kingdoms for its materials library,” she writes. "Pick a feature from an existing organism, locate its DNA, and insert it into a biological chassis. Engineered life could compute, produce energy, clean up pollution, kill pathogens, and even do the housework. Meanwhile, we’ll have to add an extra branch to the Tree of Life. The Synthetic Kingdom is part of our new nature.”
Crucial to this paradigm shift for biological reality on earth is CRISPR. In his essay A Future History of Biomedical Progress, Markov Bio’s Adam Green writes, “If sequencing and microscopy are the universal sensors of the scopes tool class, then CRISPR might be the universal genomic actuator of the scalpels tool class.”
It’s been twelve years since CRISPR made its debut in the toolkit of biologists and researchers, and already the number of specimens added to the “Synthetic Kingdom” is stunning. It has yielded tomatoes rich in Vitamin D, wheat that is resistant to mildew, and corn that is resistant to drought – and that’s only the beginning. CRISPR has also been used to modify strains of yeast to produce biofuels or other industrially-relevant compounds.
CRISPR has already been used on farm animals, where researchers tinkered with a gene that tempers muscle growth to produce breeds of super muscular cattle, pigs, sheep, and other animals. Similar experiments have also had similar effects in aquaculture, where the production of larger and more protein-packed fish and disease-resistant fish is now a possibility.
CRISPR technologies have even been explored to engineer gene drives, which cause the spread of genes throughout a species’ population. One such effort involves genetically editing mosquitoes so they are less capable of transmitting diseases such as malaria.
It has even been used to modify human embryos. In 2018, Chinese scientist He Jiankiu used CRISPR to modify the DNA of two twin girls whose father was HIV-positive. He used CRISPR to alter a protein-coding gene that would prevent the virus from entering cells. Still, the procedure was illegal and He Jiankiu was imprisoned for three years following the birth of the girls. Remarkably, it can now be said that genetically modified DNA is now officially part of humanity’s genetic lineage.
As time goes on, the technologies for performing genetic sequencing, synthesis, and engineering are becoming ever more accessible. In 2007, the cost of sequencing the human genome hovered around $1 million. Today, it costs somewhere around $600.

Source: National Human Genome Research Institute
The costs of synthesizing DNA, though not falling as precipitously as sequencing, have also come down significantly. In 2000, synthesizing a single base pair could cost up to $10, where a single gene can contain between a few hundred to two million base pairs. In 2024, the cost of synthesizing a single base pair hovered around 20 cents, and a few companies have even released benchtop machines that can allow anyone to do this on-premises.
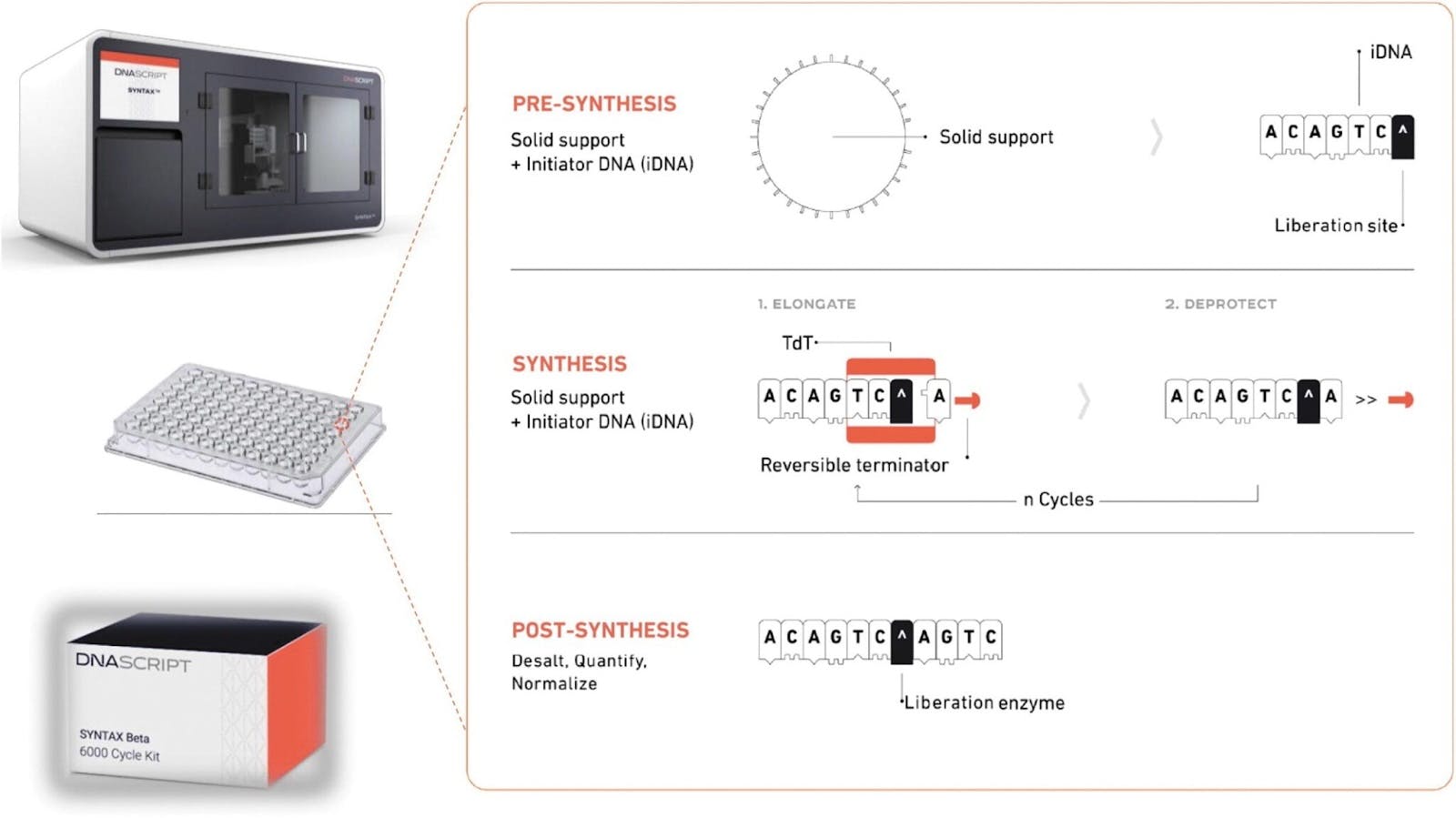
Source: GenEng News
This has led to the slow but steady emergence of a home-brew collective of biohackers, conducting advanced biological experiments in the comfort of their own home laboratories. One notable example includes Justin Atkin, who runs the YouTube account Thought Emporium. In 2018, Atkin “genetically engineered” himself to get rid of his lactose intolerance by ingesting a virus carrying some DNA that coded for lactase, an enzyme that breaks down lactose. The effect wore off after a little more than a year, but according to Atkin, greatly improved his quality of life!
Examples like this showcase some of the forthcoming dilemmas associated with widespread access to genetic technologies. Though certain regions, like the EU, require any practitioners of genetic engineering to be licensed, the same is not true in Canada, where genetic engineering only requires a license when performed on organisms with spines. The United States does not have any such practical restrictions.
This technology has already begun changing our world. The scope of its availability is just one of a myriad of factors that will need to be considered as we consider how to navigate its growing prominence.
A History of Genetic Editing
It’s a rather remarkable fact that the mechanism behind which organisms function, reproduce, and evolve on a molecular level was theorized before DNA’s role in heredity or genetics was ever confirmed. It was a theory developed by the legendary polymath, John von Neumann.
Neumann was a mathematician and engineer who had become fascinated with computers in the 1940s. In particular, he was fascinated by the mechanisms used by machines to store programs and execute their instructions. Eventually, he began to wonder if there might be parallels between the way machines operate and the way that biological organisms do.
The biggest difference between biological organisms and machines, however, was that organisms could reproduce and evolve. Naturally, this made von Neumann ask the question: what would it look like for a machine to reproduce and evolve? This line of questioning was the origin of his ‘universal self-reproducing machine’ — a theoretical device capable of evolving itself into more complex machines over generations.
This machine would have had three components: a “blueprint” describing the machine itself; a “constructor” that translates the blueprint into building instructions; and a “copying machine” that would produce a copy of those instructions. The idea, as Hashim Al-Hashimi illustrates in his article Turing, von Neumann, and the computational architecture of biological machines, is that “a machine builds a copy of itself using the instructions, then makes a copy of the instructions, and feeds them into the new machine, and so on.” This process explains how machines could reproduce, but not evolve.
To account for this, von Neumann envisioned that the copying machine would be allowed to make mistakes, leading successive generations of machines to carry mutations that, over successive generations, could produce varied and perhaps more complex versions. Von Neumann had a hunch this might be analogous to how biological organisms reproduce and evolve over time, too. As von Neumann put it as early as 1948:
“...the instruction is roughly effecting the functions of a gene. It is also clear that the copying mechanism performs the fundamental act of reproduction, the duplication of the genetic material, which is clearly the fundamental operation in the multiplication of living cells. It is also easy to see how arbitrary alterations … can exhibit certain typical traits which appear in connection with mutation, lethally as a rule, but with a possibility of continuing reproduction with a modification of traits.”
Five years later, James Watson and Francis Crick discovered the double helix structure of DNA. Seven years after that, Francis Crick posited the central dogma of molecular biology: which asserted that genetic information is stored in DNA, transcribed to RNA, and finally translated and synthesized into proteins.
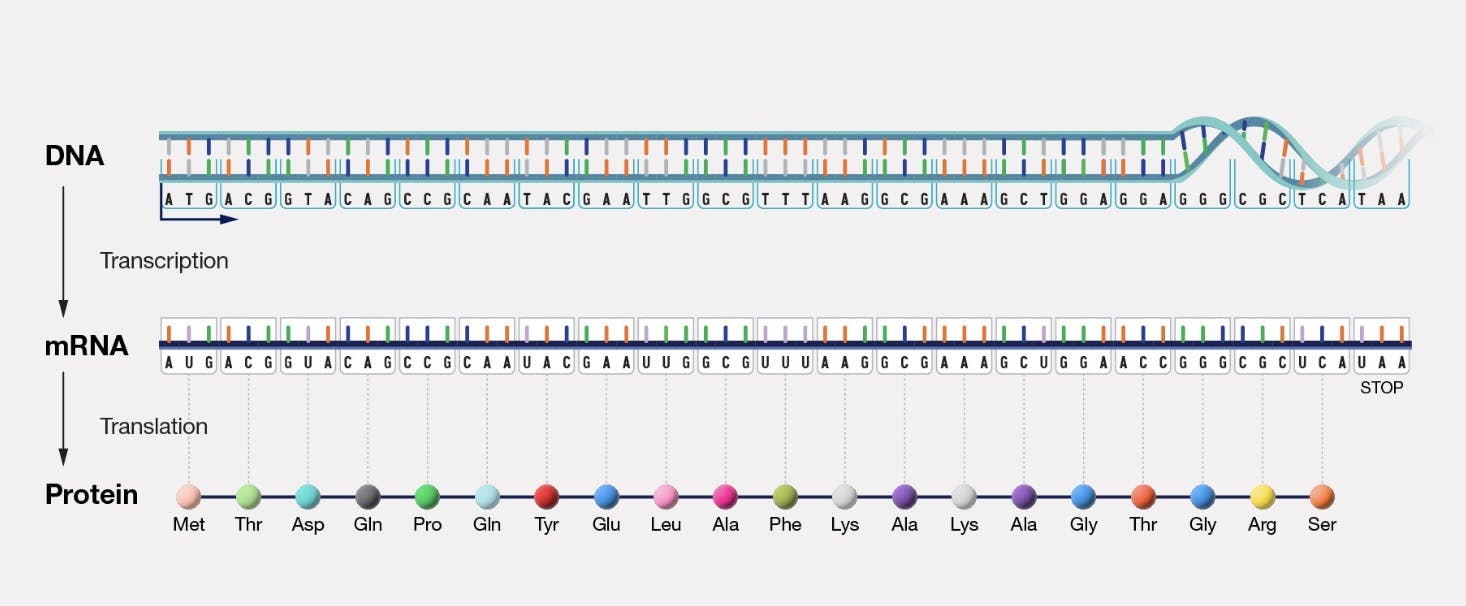
Source: National Human Genome Research Institute
This central dogma is, in fact, the core mechanism through which all biological organisms reproduce and function, and maps directly onto von Neumann’s theoretical description of a ‘universal self-reproducing machine’.
Everything seemed to be pointing to the idea that the cell was quite like a machine, albeit an extremely complicated one. Once the role of DNA was confirmed, decades of research went into understanding it in greater detail.
In the mid-1950s, critical enzymes like DNA polymerase and RNA polymerase were discovered, which were the proteins responsible for producing copies of DNA and translating DNA into RNA, respectively. These were the cells’ ‘copying machines’.
In 1965, Marshall Nirenberg cracked the genetic code, revealing how triplets of nucleotides coded for 20 possible amino acids, which when strung together could form proteins.
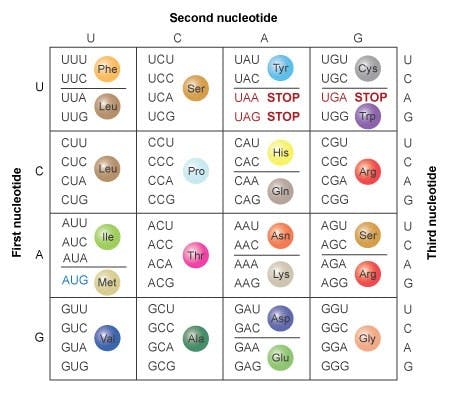
Source: Nature
Three years later, an artificial method of gene transfer was devised by Stanfield Rogers. He suggested that viruses could be instrumentalized to carry genetic material and transfer it into cells. Of course, without any methods of accurately selecting which genetic material we want to transfer, it was an idea that didn’t bear any immediate fruit.
That would change just a few years later, in 1972, when a team at UCSF and Stanford University, consisting of Stanley Cohen, Herb Boyer, and others, used enzymes to cut DNA fragments from different species and fuse the strands together to produce the first instance of recombinant DNA. Herb Boyer would go on to co-found Genentech four years later, inaugurating the biotech industry in the Bay.
Cohen and Boyer demonstrated that recombining DNA was possible, however, the method they used inserted genetic fragments into the genome at random. The search, therefore, was on for a method that could insert or remove fragments at deliberate points within the genome.
The first answer to this came in the form of ZFNs (Zinc Finger Nucleases) and TALENs (Transcription Activator-Like Effector Nucleases), which were discovered in 1996 and 2010 respectively. Both of these were enzymes, discovered in bacteria, which could be engineered to cut DNA fragments at particular junctions.
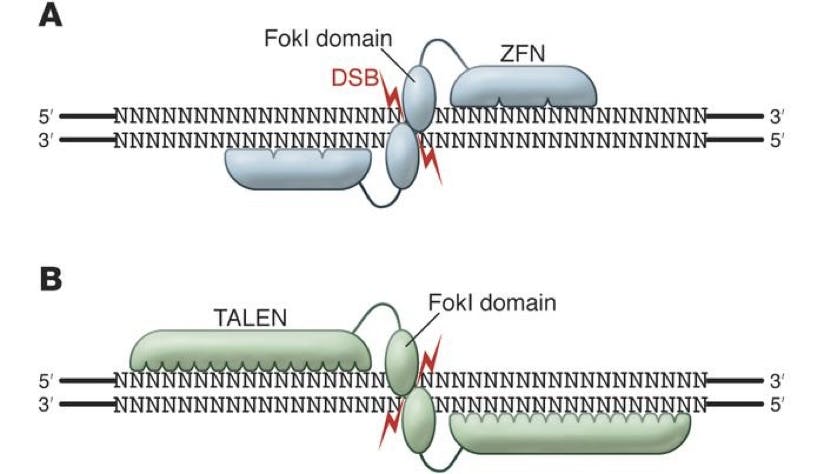
Source: The Journal of Clinical Investigation
Things were clearly getting somewhere, even if rather slowly and clunkily. Due to the nature of ZFN and TALEN technologies, both needed to be engineered to match the required cutting sequences. This took time, and the actual operation of these enzymes could be flawed. Biologists were still hoping for a tool that could deliver an even greater degree of precision for identifying the exact and unique segment of targeted DNA.
Two years after the discovery of TALENs, just such a technology was introduced to the world in the form of CRISPR. CRISPR did not arise from the technologies that preceded it. Its story began with the work of Japanese scientist Yoshizumi Ishino, who, in 1987, was studying E. coli. He noticed something strange in the bacteria’s genome, namely identical sections of repeated code, interspersed by unique encodings of ‘spacer’ DNA. These strange genetic sections puzzled him, and it would take over a decade for researchers to get any clarity on what they meant.

Source: Explore Biology
A Spanish researcher, Francisco Mojica, finally named these odd sequences CRISPRs, in the early 2000s, short for clustered regularly interspaced short palindromic repeats. Around that time researchers realized that the DNA in the spacer segments mapped perfectly onto the DNA of known bacteriophages, or viruses. This led Mojica to posit that these CRISPRs must work as a kind of memory, part of a larger bacterial immune system.
Soon after, CRISPR-associated sequence or “Cas" genes were identified right next to the strips of CRISPR code. These Cas genes coded for helicase and nuclease enzymes, responsible for unwinding and cutting DNA segments.
By 2007, due to research performed by Rodolphe Barrangou, the core idea became clear. Bacteria would store viral DNA within their own genome as spacer sequences, and later on, this stored information was used as a reference to destroy viral DNA should it ever be encountered again.
As it turned out, hidden inside an E. coli cell was the exact suite of precision tools biologists had been looking for. However, it wasn’t an easy road to convincing funding organizations or scientific publications that research into the immune system of bacteria was worth it. The research that led Barrangou to confirm the immune function of bacteria was conducted through the cooperation of the yogurt company Danisco, but as Barrangou mentions, “It took … 6 months to convince the management at Danisco that our results were worth publishing. It also was not trivial to convince Science magazine and its reviewers to have interest in publishing a study on a peculiar genetic locus from a yogurt starter culture. But eventually, the paper was submitted in 2006 and published in 2007.”
Once Barrangou published, however, the implications and applications of this mechanism became clear. The only remaining questions were figuring out the exact mechanisms by which the CRISPR-Cas system worked.
Ultimately, the complete puzzle was solved by Jennifer Doudna and Emmanuelle Charpentier in 2012. They discovered that there were three components involved — the Cas protein capable of cutting DNA, a tracrRNA strand that helped guide the Cas protein to the appropriate location, and a crRNA strand containing the appropriate DNA sequence to target.

Source: Explore Biology
Doudna and Charpentier realized that CRISPR had all the components required to be a general-purpose gene editing technology. Doudna and Charpentier also realized that the tracrRNA and crRNA strands could be united into one — requiring the synthesis of just one strand of RNA to guide the Cas enzyme to the right location.
The system that Doudna and Charpentier exposed was far more effective than any gene editing technology that came before. It could work in any species, in vivo — in living organisms — just as well as it worked in a test tube. And, it had many less moving pieces and components that needed to be synthesized to work, unlike ZFNs and TALENs, which needed two distinct types of proteins. The CRISPR-Cas complex required the synthesis of just one strand of guide RNA. Due to the system’s ease of use, speed, and high precision, it became the de facto state of the art in gene editing techniques and won Doudna and Charpentier the Nobel Prize in 2020.
Outstanding Challenges and Future Directions
One of the most notable features of the CRISPR system is its incredible flexibility. By tweaking the Cas enzyme and guide RNA used, everything from knocking out the function of genes to introducing entirely new genetic traits became possible. CRISPR systems can even be used to perform multiplex editing — which means performing edits on a number of genes at the same time. Alternatively, they can be used to edit just a single nucleotide. It’s these abilities, capable of being carried out in virtually any organism, that have led to the explosion of interest and research employing CRISPR.
With that said, there are still many technical hurdles for CRISPR editing technologies to overcome. While CRISPR is much more precise than other methods, it is still a biological mechanism that can be prone to error. While the CRISPR-Cas complex can target the desired DNA location precisely, it does not guarantee that the edited result will come out exactly as expected.
In CRISPR’s early days, the chances of off-target cuts happening were common. With time, these kinds of issues were mitigated by improving on the design of guide RNA sequences. Design of an effective guide RNA limited the probability of the Cas enzyme confusing the desired section for another within the genome.
Another outstanding technical challenge is managing what happens once the DNA is cut. The original CRISPR system relied on inherent DNA repair mechanisms like NHEJ (Non-homologous end joining), which can at times introduce scar tissue or unwanted, random insertions that impede the correct performance of the edited genes.
Ensuring proper DNA repair post-editing continues to be a challenge researchers are working to perfect. However, there are still many hurdles. For instance, in 2020, one study demonstrated that a CRISPR-Cas experiment to repair a genetic mutation that causes hereditary mutation succeeded in only 50% of the cells. The other half of the time, the DNA repair failed so drastically that affected cells discarded the entire chromosome where the error resided.
Since 2016, a number of techniques, like prime editing, have been developed to prevent causing large breaks that might lead natural mechanisms like NHEJ to introduce unwanted material to the genome. Additionally, a number of “second-generation” CRISPR systems are being investigated which can perform both the editing and DNA repair steps to retrain more control over the entire process.
Yet another challenge is the problem of actually delivering this CRISPR-Cas complex to desired cells where the genetic change is to be made. Thus far, researchers have enjoyed great success by genetically editing cells ex vivo and then re-inserting them into patients. This has worked particularly well with CAR-T technologies, like those which target B-cells, rather than any kind of structured tissue. It is in vivo operations that are particularly difficult to engineer correctly.
Presently, the best approach to deliver CRISPR-Cas complexes to the correct cell involves using viral vectors that target the desired cells. However, in many cases, the required CRISPR-Cas complex is simply too big for the size of the virus. With more sophisticated CRISPR editing tools, like prime editing, a virus simply isn’t big enough to fit it all.
It is outstanding challenges like this that make existing genetic therapies on the market prohibitively expensive. The entire lifecycle of developing and producing gene therapies is itself slow and expensive, which contributes to the high and rising costs. Designing, synthesizing, and iterating on numerous guide RNAs to test the performance of certain therapies can reach exorbitant levels. The average cost of a gene therapy is between $1 million and $2 million, while the average cost of bringing a gene therapy to market falls just under $2 billion.
This is why a number of companies are building a whole new industry designed to address exactly these process bottlenecks. Biomanufacturing firms like Synthego are attempting to be a one-stop-shop for all labs researching new gene therapies or any other applications that involve genetic editing tools.
Uniting the software design of the guide RNA, handling its manufacture, its insertion into targeted cells, and verifying the effectiveness of the edits made is the full-service offering provided by companies like this.

Source: Axial
Full-stack biomanufacturing shops draw on a number of technologies to perform all of these tasks as accurately and affordably as possible. It’s here that the intimate relationships between DNA sequencing, DNA synthesis technologies, and the myriad of CRISPR techniques emerge. It goes to show how technical improvements in one area, like falling DNA synthesis costs, can have massive implications for the scalability of editing techniques like CRISPR. The possibilities afforded by gene therapies are immense, including everything from treating auto-immune diseases to various types of cancers and much more. Though CRISPR technology is still relatively young, part of the road to perfecting it will need to involve building out better infrastructure and tools to iterate on its improvement more rapidly. Bringing down the costs of experimentation will allow researchers to iterate more rapidly and offer more affordable therapies down the line.
Modern synthetic biology and genetic engineering is a concerted effort — relying on incremental discoveries from researchers, process advancements across a host of genetic technologies, and novel insights from the realm of computational biology. Though CRISPR is a crucial technology that revolutionized the field of biology in the 21st century, it is only one piece of a larger collective that works in concert to expand the horizons of biology.
Disclosure: Nothing presented within this article is intended to constitute legal, business, investment or tax advice, and under no circumstances should any information provided herein be used or considered as an offer to sell or a solicitation of an offer to buy an interest in any investment fund managed by Contrary LLC (“Contrary”) nor does such information constitute an offer to provide investment advisory services. Information provided reflects Contrary’s views as of a time, whereby such views are subject to change at any point and Contrary shall not be obligated to provide notice of any change. Companies mentioned in this article may be a representative sample of portfolio companies in which Contrary has invested in which the author believes such companies fit the objective criteria stated in commentary, which do not reflect all investments made by Contrary. No assumptions should be made that investments listed above were or will be profitable. Due to various risks and uncertainties, actual events, results or the actual experience may differ materially from those reflected or contemplated in these statements. Nothing contained in this article may be relied upon as a guarantee or assurance as to the future success of any particular company. Past performance is not indicative of future results. A list of investments made by Contrary (excluding investments for which the issuer has not provided permission for Contrary to disclose publicly, Fund of Fund investments and investments in which total invested capital is no more than $50,000) is available at www.contrary.com/investments.
Certain information contained in here has been obtained from third-party sources, including from portfolio companies of funds managed by Contrary. While taken from sources believed to be reliable, Contrary has not independently verified such information and makes no representations about the enduring accuracy of the information or its appropriateness for a given situation. Charts and graphs provided within are for informational purposes solely and should not be relied upon when making any investment decision. Please see www.contrary.com/legal for additional important information.